Code
library (tidyverse)library (palmerpenguins)library (showtext)library (car)font_add_google ("Lato" , "Lato" )showtext_auto ()library (patchwork)library (ggeffects)library (performance)library (broom)library (flextable)library (DHARMa)library (GGally)library (MuMIn)
multiple linear regression equation
\[
\begin{align}
y &= \beta_0 + \beta_1x_1 + \beta_2x_2 + ... \beta_kx_k + \epsilon
\end{align}
\]
plant example
Code
set.seed (666 )# sample size <- 64 <- tibble (# predictor variables temperature = round (rnorm (n = n, mean = 28 , sd = 1 ), digits = 1 ),light = round (rnorm (n = n, mean = 1 , sd = 0.2 ), digits = 1 ),ph = rnorm (n = n, mean = 7 , sd = 0.01 ),# response: growth in cm/week growth = light* rnorm (n = n, mean = 0.3 , sd = 0.1 ) + temperature/ round (rnorm (n = n, mean = 5 , sd = 0.1 ))
Code
<- lm (growth ~ light + temperature + ph, data = plant_df)
Code
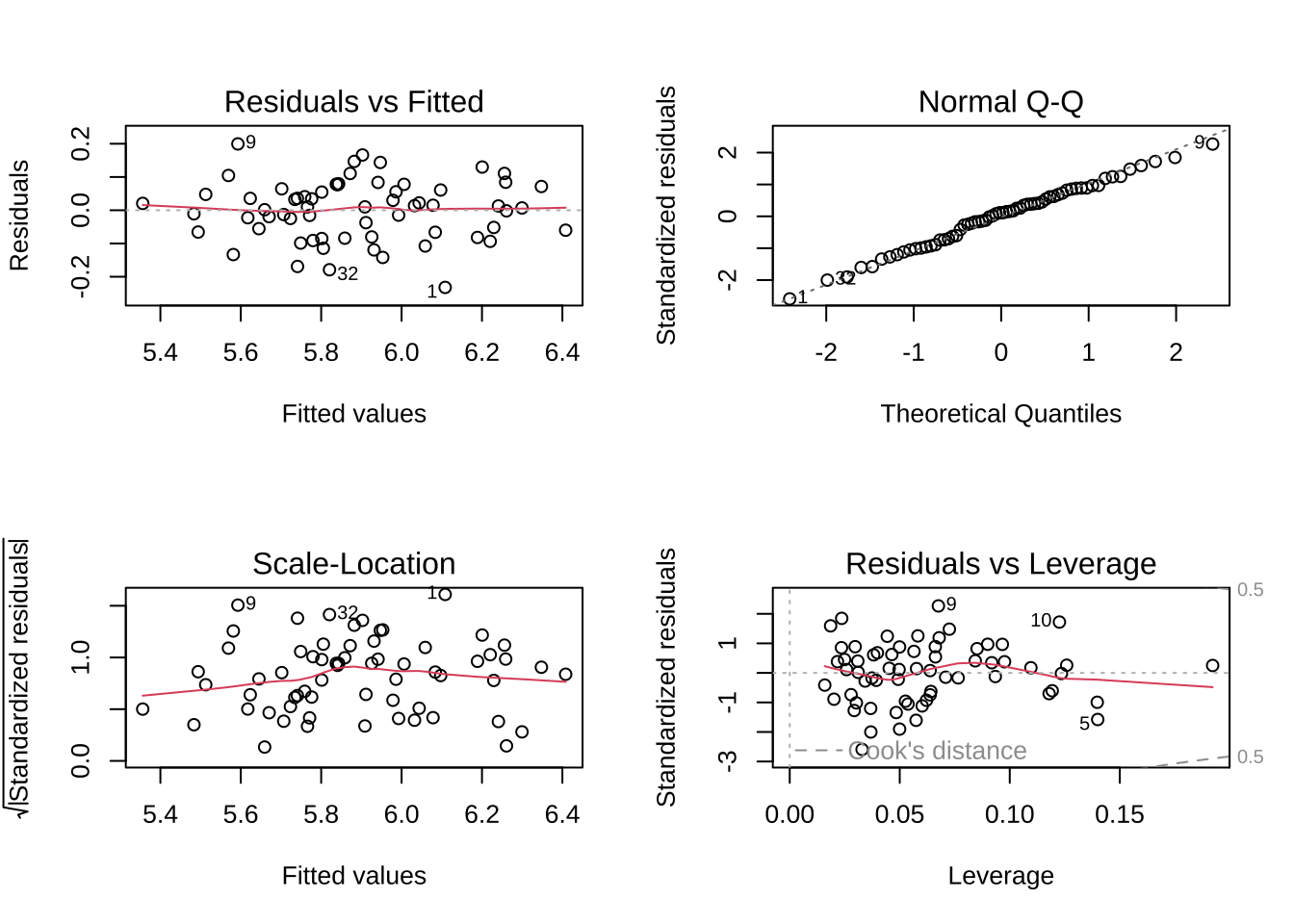
par (mfrow = c (2 , 2 ))plot (plant_model)
Code
Code
pairs (plant_df, upper.panel = NULL )
Code
Code
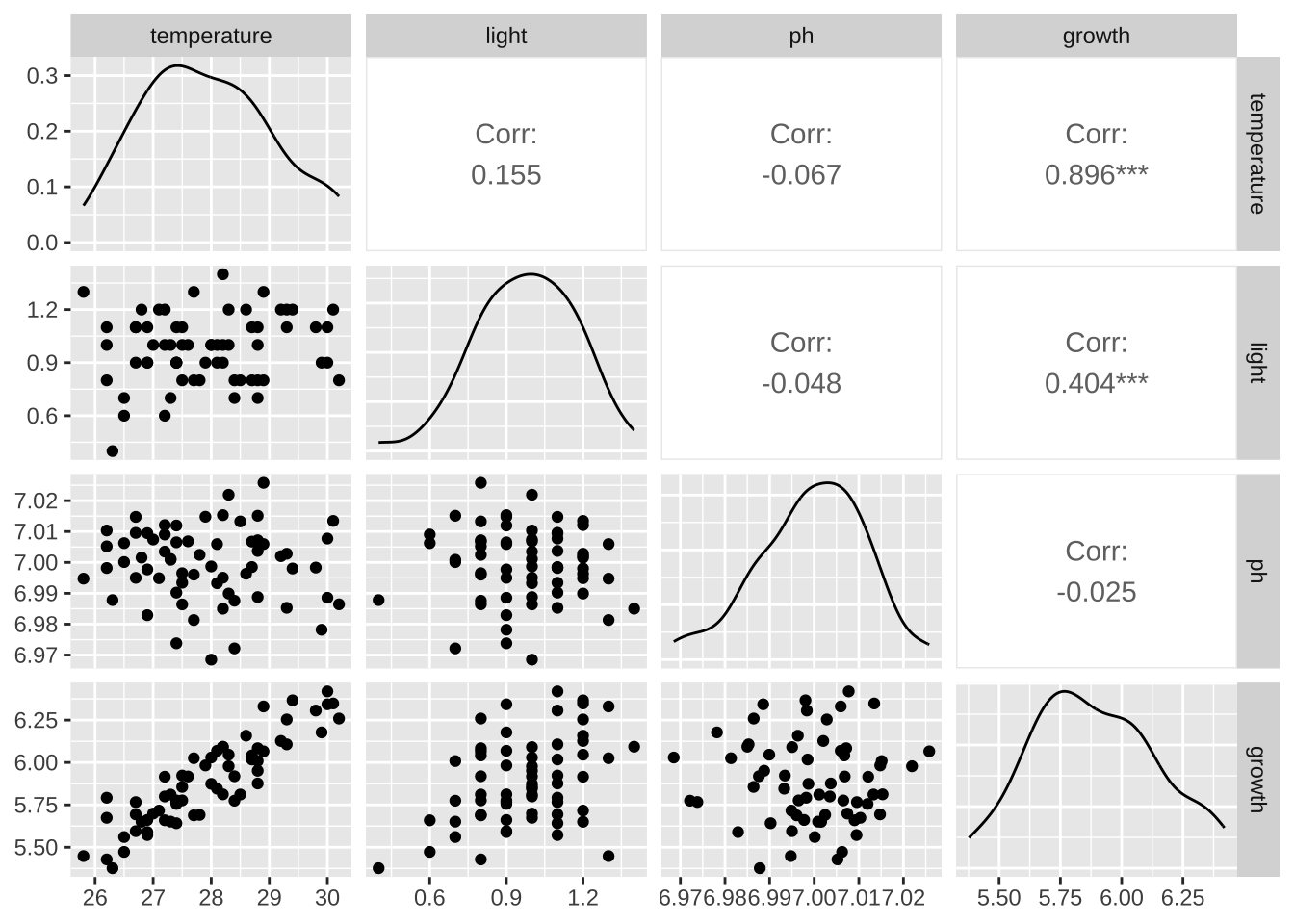
temperature light ph growth
temperature 1.00000000 0.15540780 -0.06657697 0.89551840
light 0.15540780 1.00000000 -0.04769987 0.40397013
ph -0.06657697 -0.04769987 1.00000000 -0.02466729
growth 0.89551840 0.40397013 -0.02466729 1.00000000
Code
light temperature ph
1.026223 1.028447 1.005897
Code
Call:
lm(formula = growth ~ light + temperature + ph, data = plant_df)
Residuals:
Min 1Q Median 3Q Max
-0.23209 -0.06571 0.01010 0.06173 0.19950
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -6.67140 6.80194 -0.981 0.331
light 0.35196 0.05939 5.926 1.63e-07 ***
temperature 0.19626 0.01058 18.558 < 2e-16 ***
ph 0.96241 0.96806 0.994 0.324
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.0911 on 60 degrees of freedom
Multiple R-squared: 0.8759, Adjusted R-squared: 0.8696
F-statistic: 141.1 on 3 and 60 DF, p-value: < 2.2e-16
Code
Analysis of Variance Table
Response: growth
Df Sum Sq Mean Sq F value Pr(>F)
light 1 0.65459 0.65459 78.8700 1.567e-12 ***
temperature 1 2.85039 2.85039 343.4377 < 2.2e-16 ***
ph 1 0.00820 0.00820 0.9884 0.3241
Residuals 60 0.49797 0.00830
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
For example, temperature F-value:
\[
\begin{align}
F &= \frac{2.85039}{0.00830} \\
&= 343.4205
\end{align}
\]
frog example
generating data
Code
set.seed (666 )<- 87 <- cbind (# predictor variables color = sample (x = c ("blue" , "green" , "red" ), size = frog_n, replace = TRUE , prob = c (0.3 , 0.3 , 0.3 )),weight = (round (rnorm (n = frog_n, mean = 3 , sd = 0.3 ), 2 )),pattern = sample (x = c ("striped" , "spotted" , "none" ), size = frog_n, replace = TRUE , prob = c (0.3 , 0.3 , 0.3 ))%>% as_tibble () %>% mutate (weight = as.numeric (weight),color = as.factor (color),pattern = as.factor (pattern)) %>% group_by (color, pattern) %>% # response variable mutate (toxicity = case_when (== "blue" & pattern == "striped" ~ rnorm (n = length (color), mean = 5 , sd = 1 ),== "blue" & pattern == "spotted" ~ rnorm (n = length (color), mean = 4 , sd = 1 ),== "green" & pattern == "striped" ~ rnorm (n = length (color), mean = 4 , sd = 1 ),== "green" & pattern == "spotted" ~ rnorm (n = length (color), mean = 3 , sd = 1 ),== "red" ~ rnorm (n = length (color), mean = 6 , sd = 1 ),TRUE ~ rnorm (n = length (color), mean = 2 , sd = 1 )%>% ungroup ()
plotting data
Code
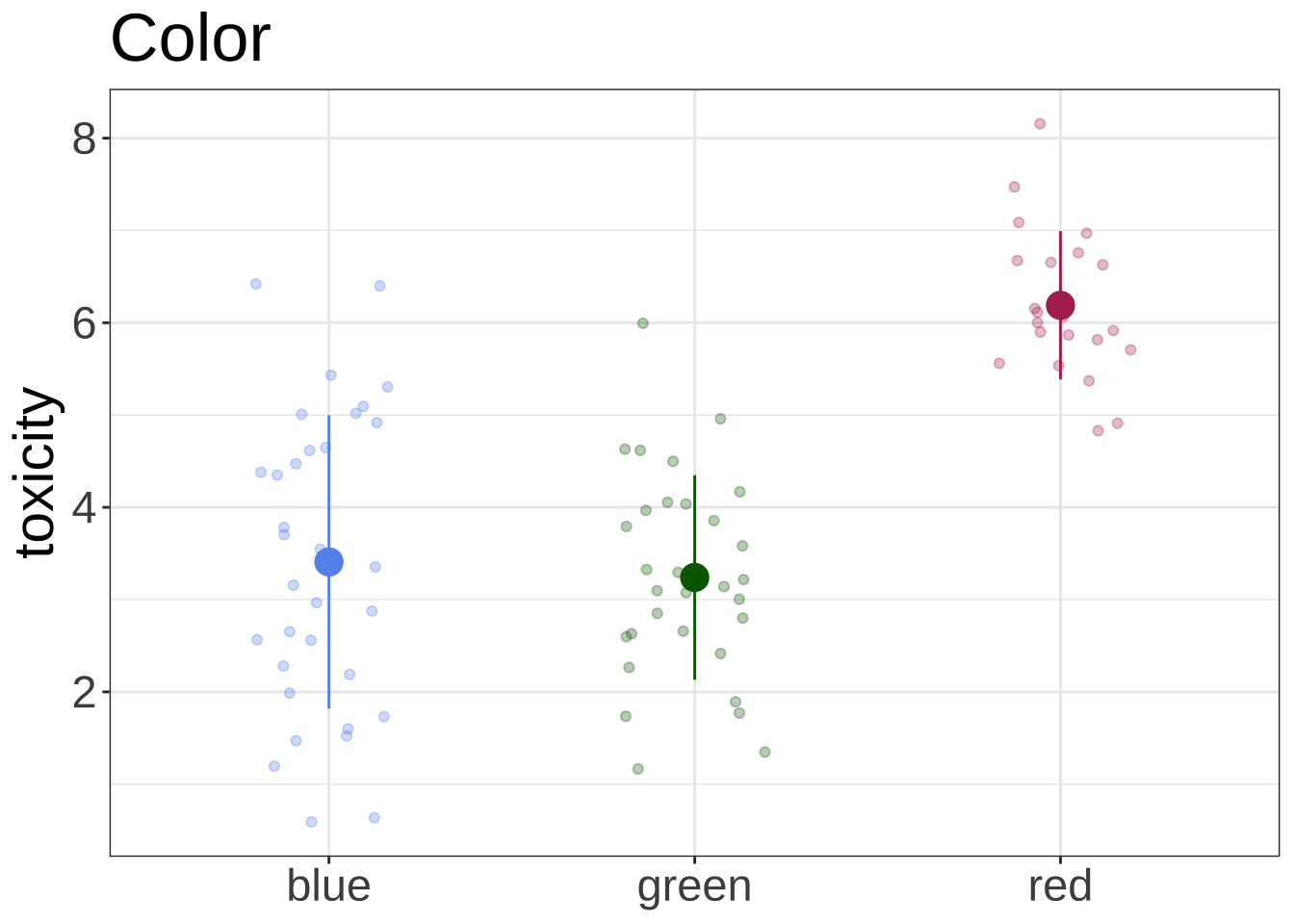
<- "cornflowerblue" <- "darkgreen" <- "maroon" <- "grey1" <- "grey50" <- "grey80" ggplot (data = df, aes (x = color, y = toxicity, color = color, fill = color)) + geom_jitter (width = 0.2 , height = 0 , alpha = 0.3 ) + scale_color_manual (values = c ("blue" = blue_col, "green" = green_col, "red" = red_col)) + scale_fill_manual (values = c ("blue" = blue_col, "green" = green_col, "red" = red_col)) + stat_summary (geom = "pointrange" , fun = mean, fun.min = function (x) mean (x) - sd (x), fun.max = function (x) mean (x) + sd (x), shape = 21 , size = 1 ) + # geom_point(position = position_jitter(width = 0.2, height = 0, seed = 666), alpha = 0.3) + labs (title = "Color" ) + theme_bw () + theme (legend.position = "none" ,axis.title.x = element_blank (),text = element_text (size = 22 ))
Code
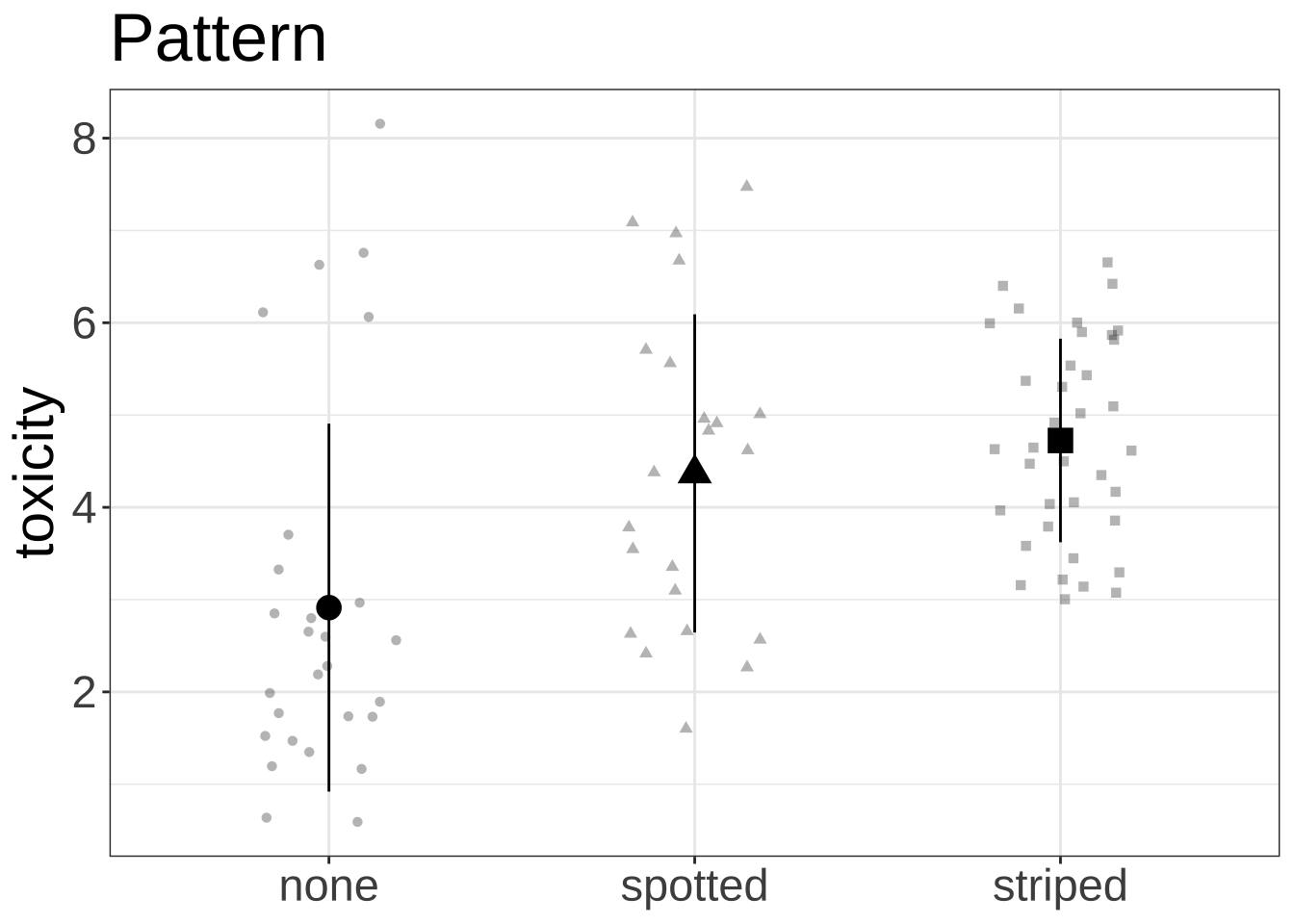
ggplot (data = df, aes (x = pattern, y = toxicity, shape = pattern)) + geom_jitter (width = 0.2 , height = 0 , alpha = 0.3 ) + # scale_color_manual(values = c("blue" = blue_col, "green" = green_col, "red" = red_col)) + # scale_fill_manual(values = c("striped" = striped_col, "spotted" = spotted_col, "none" = none_col)) + stat_summary (geom = "pointrange" , fun = mean, fun.min = function (x) mean (x) - sd (x), fun.max = function (x) mean (x) + sd (x), size = 1 ) + labs (title = "Pattern" ) + theme_bw () + theme (legend.position = "none" ,axis.title.x = element_blank (),text = element_text (size = 22 ))
Code
ggplot (data = df, aes (x = weight, y = toxicity)) + geom_point () + # geom_smooth(method = "lm") + labs (title = "Weight" ) + theme_bw () + theme (legend.position = "none" ,axis.title.x = element_blank (),text = element_text (size = 22 ))
Code
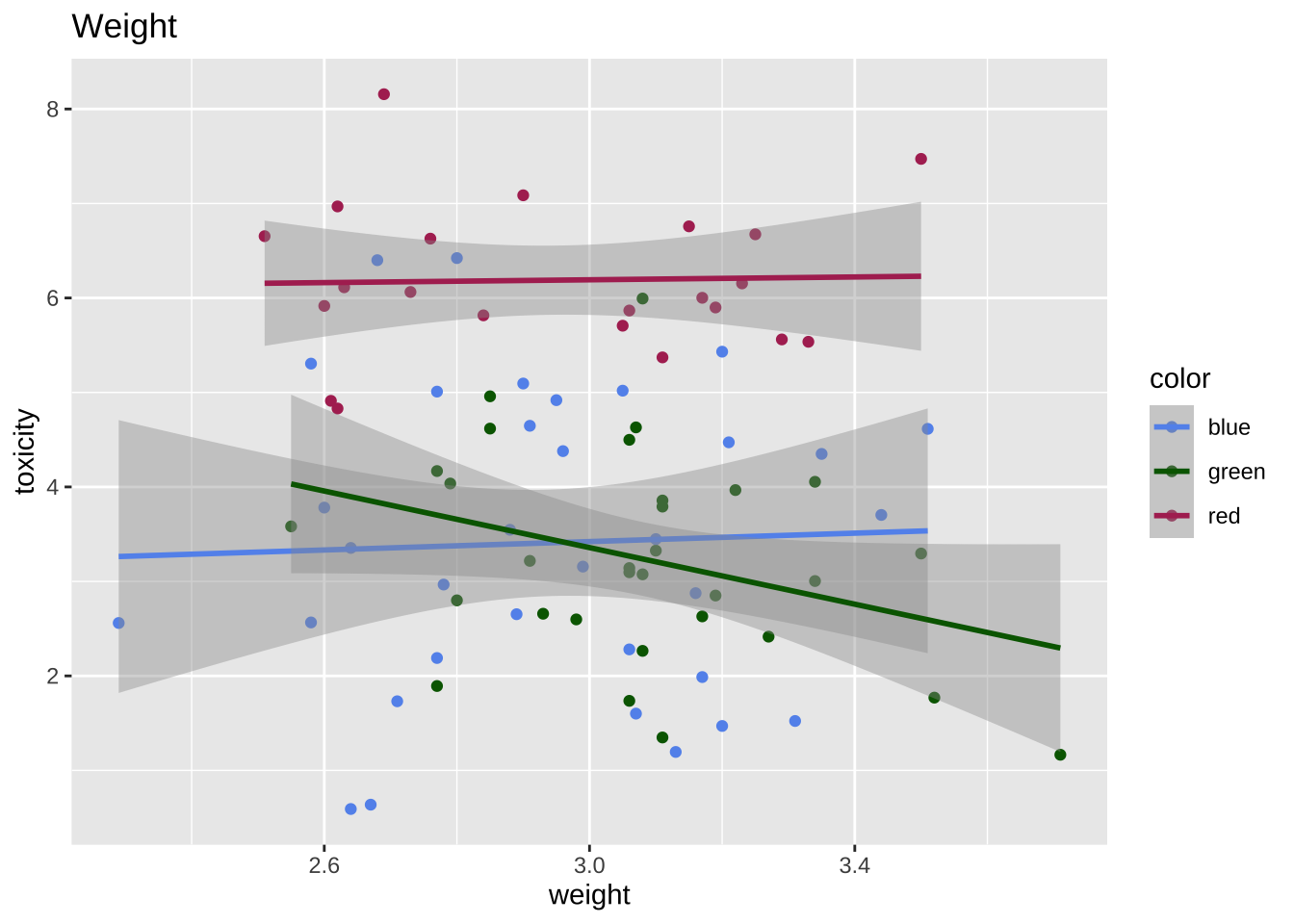
ggplot (data = df, aes (x = weight, y = toxicity, color = color)) + geom_point () + scale_color_manual (values = c ("blue" = blue_col, "green" = green_col, "red" = red_col)) + geom_smooth (method = "lm" ) + labs (title = "Weight" )
model
Code
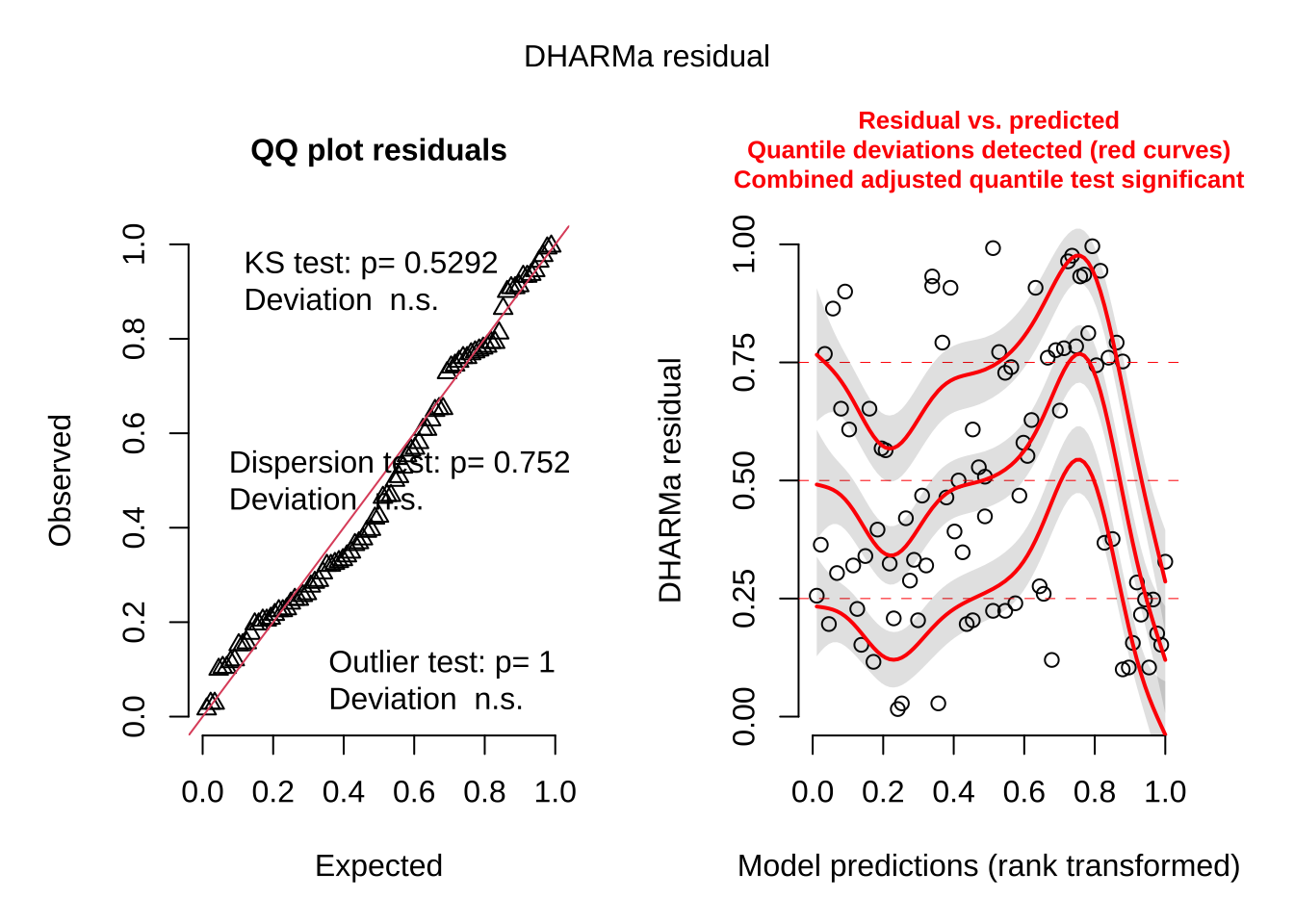
<- lm (toxicity ~ weight + color + pattern, data = df)simulateResiduals (model, plot = TRUE )
Object of Class DHARMa with simulated residuals based on 250 simulations with refit = FALSE . See ?DHARMa::simulateResiduals for help.
Scaled residual values: 0.208 0.608 0.196 0.468 0.744 0.792 0.908 0.792 0.992 0.256 0.324 0.768 0.468 0.32 0.204 0.908 0.288 0.76 0.156 0.104 ...
diagnostics
Code
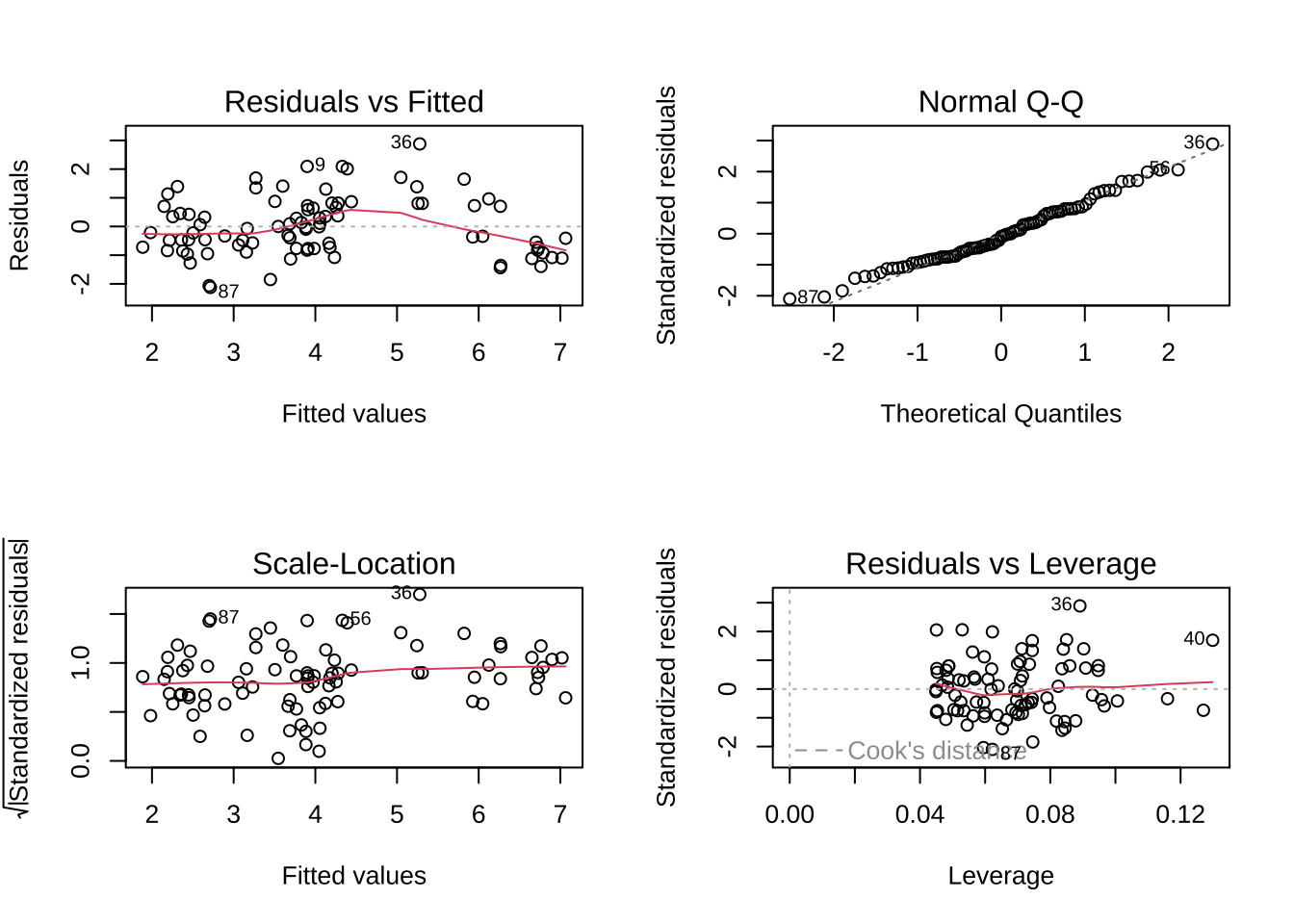
par (mfrow = c (2 , 2 ))plot (model)
model summary
Code
# F-statistic: 31.05 on 5 and 81 DF, p-value: < 2.2e-16 # total SSE - SSE of residuals divided by degrees of freedom <- 6.932+140.09+21.63 <- 1 + 2 + 2 <- 88 <- (totalSSE/ 5 )/ (errorSSE/ 81 )
Code
# for a single coefficient <- 6.932 <- 1 <- 1.086 <- (weightMS/ weightdf)/ errorMS
Code
<- 70.045 <- 2 <- (colorMS/ colordf)/ errorMS
Code
# residual mean sq = 1.086 (denominator) # equation: t = 5.5 - 0.74*W - 0.97*green + 2.1*red + 0.85*spotted + 1.2*striped
Code
<- summary (model)Anova (model)
Anova Table (Type II tests)
Response: toxicity
Sum Sq Df F value Pr(>F)
weight 1.547 1 1.4194 0.237
color 119.288 2 54.7107 9.229e-16 ***
pattern 44.843 2 20.5669 5.981e-08 ***
Residuals 88.304 81
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Code
Model selection table
(Intrc) color pttrn weght df logLik AICc delta
model 4.045 + + 1 7 -124.095 263.6 0
Models ranked by AICc(x)
\[
\hat{y}_h \pm t_{(1-\alpha/2, n-2)}*\sqrt{MSE*(\frac{1}{n}+\frac{(x_h-\bar{x})^2}{\sum(x_i-\bar{x})^2})}
\]
\[
MSE = \frac{\sum(y_i-\hat{y})^2}{n}
\]
Code
tidy (model, conf.int = TRUE , conf.level = 0.95 )
# A tibble: 6 × 7
term estimate std.error statistic p.value conf.low conf.high
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 (Intercept) 4.04 1.26 3.21 1.92e- 3 1.54 6.55
2 weight -0.504 0.423 -1.19 2.37e- 1 -1.35 0.338
3 colorgreen -0.289 0.267 -1.08 2.82e- 1 -0.821 0.243
4 colorred 2.59 0.290 8.91 1.17e-13 2.01 3.17
5 patternspotted 0.952 0.303 3.14 2.34e- 3 0.349 1.55
6 patternstriped 1.70 0.264 6.41 8.92e- 9 1.17 2.22
Code
c ("lower" = model_summary$ coef[2 ,1 ] - qt (0.975 , df = model_summary$ df[2 ]) * model_summary$ coef[2 , 2 ],"upper" = model_summary$ coef[2 ,1 ] + qt (0.975 , df = model_summary$ df[2 ]) * model_summary$ coef[2 , 2 ])
lower upper
-1.3451604 0.3375674
Confidence interval for a single coefficient: in words: estimate plus or minus the t-value at your confidence level * standard error
Citation BibTeX citation:
@online{bui2023,
author = {Bui, An},
title = {Lecture 08 Figures},
date = {2023-05-22},
url = {https://an-bui.github.io/ES-193DS-W23/lecture/lecture-08_2023-05-22.html},
langid = {en}
}
For attribution, please cite this work as: