Code
# should haves
library(tidyverse)
library(here)
library(lterdatasampler)
# would be nice to have
library(performance)
library(broom)
library(flextable)
library(ggeffects)
library(car)May 17, 2023
How does stem length predict stem dry mass?
Look at your data (we’ve looked at this data set before, so we won’t go through it now)
Then, create some exploratory data visualization:
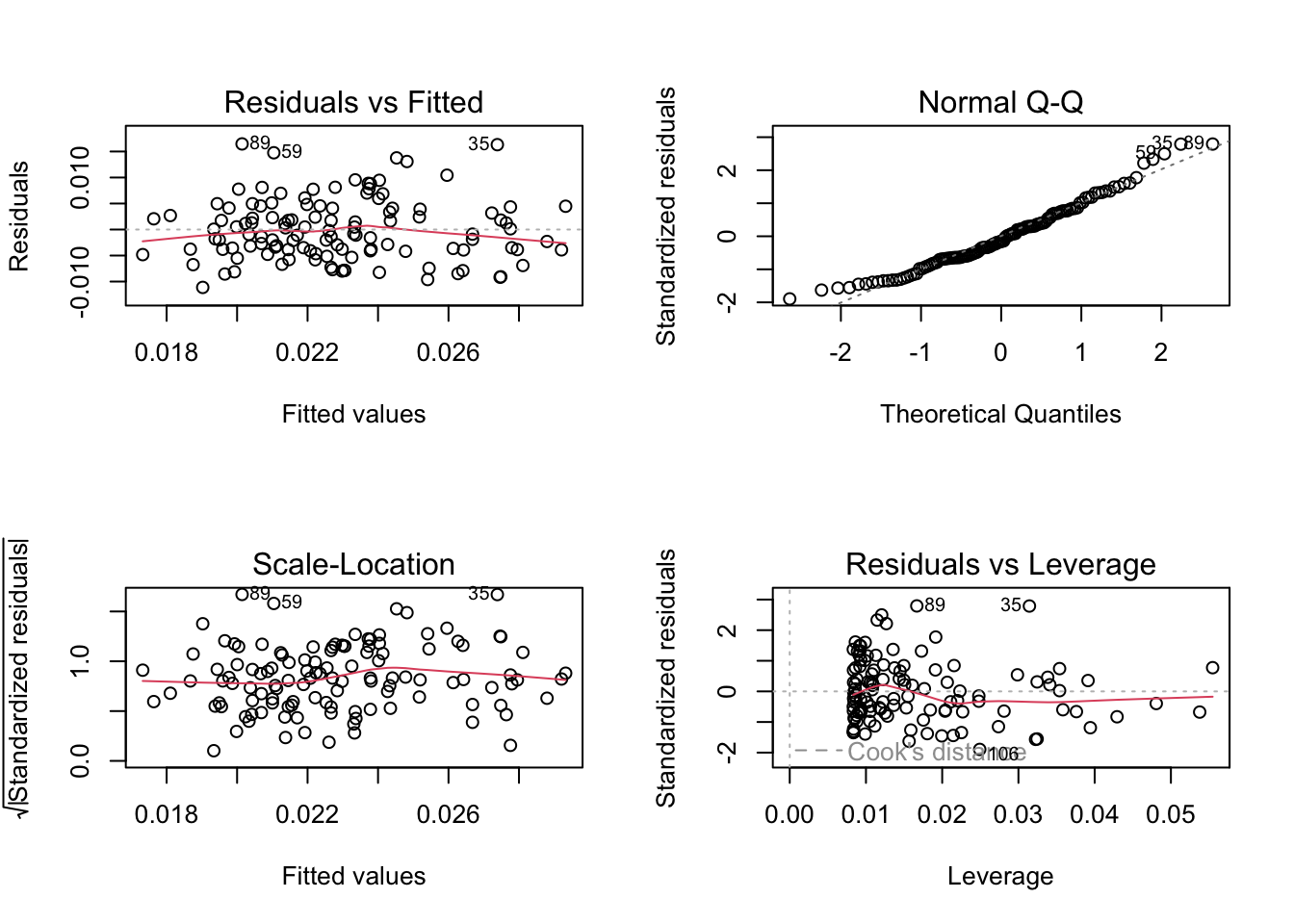
Seems like there should be a relationship between dry mass and length! Let’s try a model:
# Predicted values of stem_dry_mass
stem_length | Predicted | 95% CI
--------------------------------------
50 | 0.02 | [0.01, 0.02]
60 | 0.02 | [0.02, 0.02]
70 | 0.02 | [0.02, 0.02]
80 | 0.02 | [0.02, 0.02]
90 | 0.02 | [0.02, 0.03]
100 | 0.03 | [0.02, 0.03]
110 | 0.03 | [0.03, 0.03]
120 | 0.03 | [0.03, 0.03]plot_predictions <- ggplot(data = maples_data,
aes(x = stem_length, y = stem_dry_mass)) +
# first plot the underlying data from maples_data
geom_point() +
# then plot the predictions
geom_line(data = predictions,
aes(x = x, y = predicted),
color = "blue", linewidth = 1) +
# then plot the 95% confidence interval from ggpredict
geom_ribbon(data = predictions,
aes(x = x, y = predicted, ymin = conf.low, ymax = conf.high),
alpha = 0.2) +
# theme and meaningful labels
theme_bw() +
labs(x = "Stem length (mm)",
y = "Stem dry mass (g)")
plot_predictions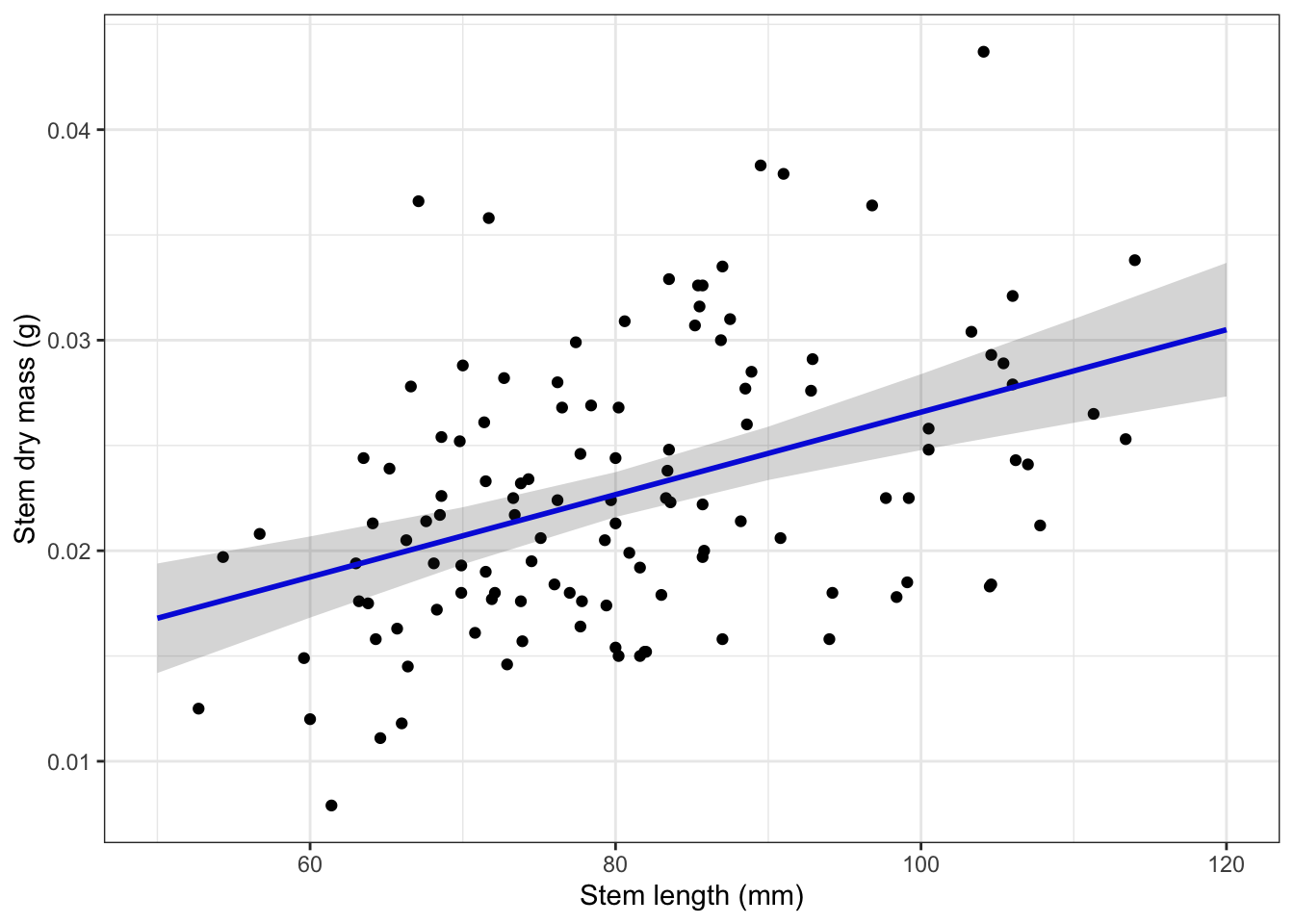
Call:
lm(formula = stem_dry_mass ~ stem_length, data = maples_data)
Residuals:
Min 1Q Median 3Q Max
-0.0111253 -0.0039117 -0.0009091 0.0040911 0.0164587
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 7.003e-03 3.212e-03 2.180 0.0312 *
stem_length 1.958e-04 3.909e-05 5.009 1.94e-06 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.005944 on 118 degrees of freedom
Multiple R-squared: 0.1753, Adjusted R-squared: 0.1683
F-statistic: 25.09 on 1 and 118 DF, p-value: 1.94e-06Analysis of Variance Table
Response: stem_dry_mass
Df Sum Sq Mean Sq F value Pr(>F)
stem_length 1 0.0008864 0.00088642 25.089 1.94e-06 ***
Residuals 118 0.0041691 0.00003533
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1model summary table:
# don't name this chunk! some intricacies with Quarto: do not name chunks with tables in them
model_squares_table <- tidy(model_squares) %>%
# round the sum of squares and mean squares columns to have 5 digits (could be less)
mutate(across(sumsq:meansq, ~ round(.x, digits = 5))) %>%
# round the F-statistic to have 1 digit
mutate(statistic = round(statistic, digits = 1)) %>%
# replace the very very very small p value with < 0.001
mutate(p.value = case_when(
p.value < 0.001 ~ "< 0.001"
)) %>%
# rename the stem_length cell to be meaningful
mutate(term = case_when(
term == "stem_length" ~ "Stem length (mm)",
TRUE ~ term
)) %>%
# make the data frame a flextable object
flextable() %>%
# change the header labels to be meaningful
set_header_labels(df = "Degrees of Freedom",
sumsq = "Sum of squares",
meansq = "Mean squares",
statistic = "F-statistic",
p.value = "p-value")
model_squares_tableterm | Degrees of Freedom | Sum of squares | Mean squares | F-statistic | p-value |
|---|---|---|---|---|---|
Stem length (mm) | 1 | 0.00089 | 0.00089 | 25.1 | < 0.001 |
Residuals | 118 | 0.00417 | 0.00004 |
Note! We didn’t get to analysis of variance in workshop on Wednesday. We will do it next week.
Do coastal giant salamander lengths differ by units?
sal <- and_vertebrates %>%
# filter for the species and unit type
filter(species == "Coastal giant salamander",
unittype %in% c("C", "P", "SC")) %>%
# creating a new column with the full unit name
mutate(unit_name = case_when(
unittype == "C" ~ "cascade",
unittype == "P" ~ "pool",
unittype == "SC" ~ "channel"
)) %>%
# transforming the length variable with a natural log
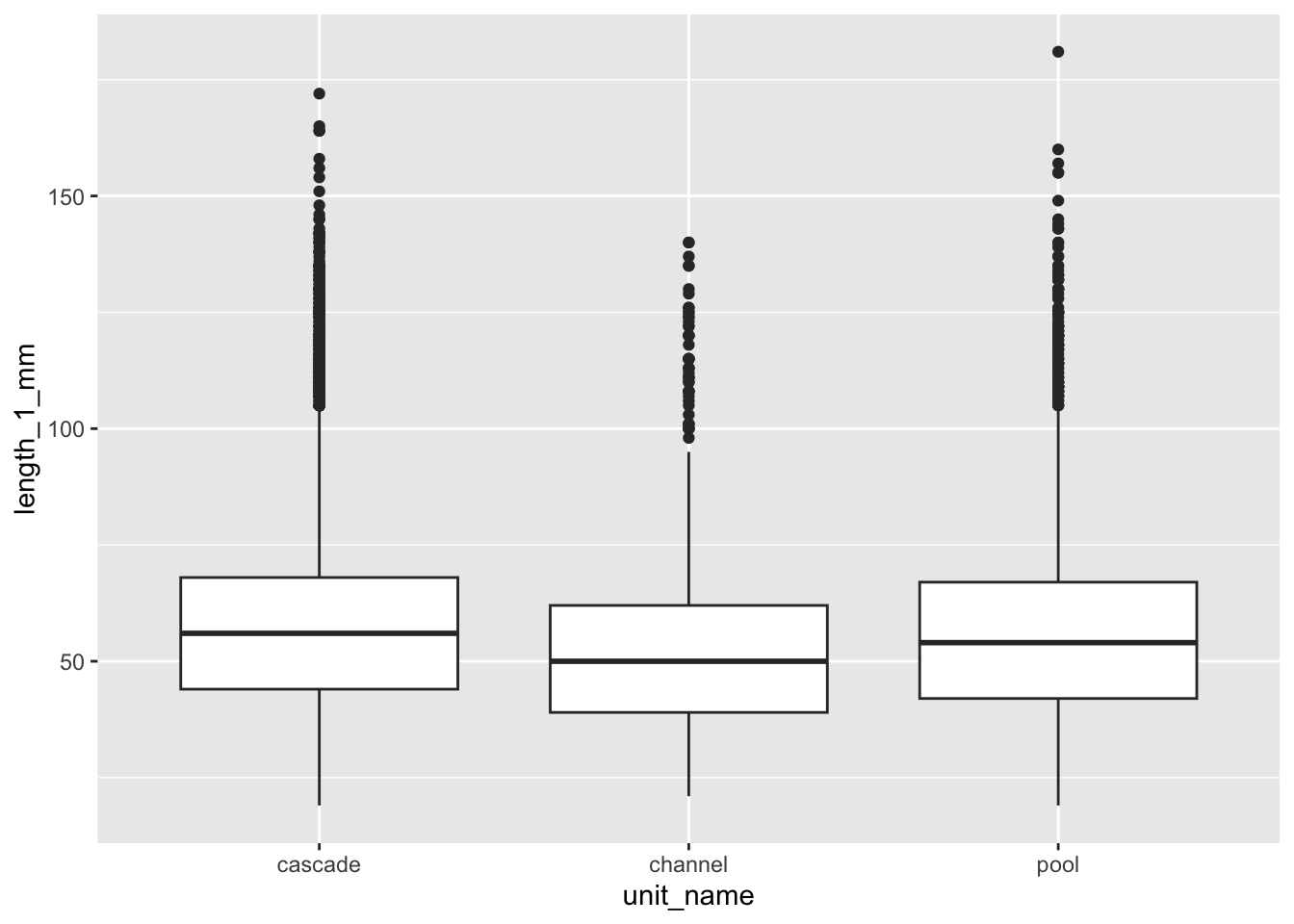
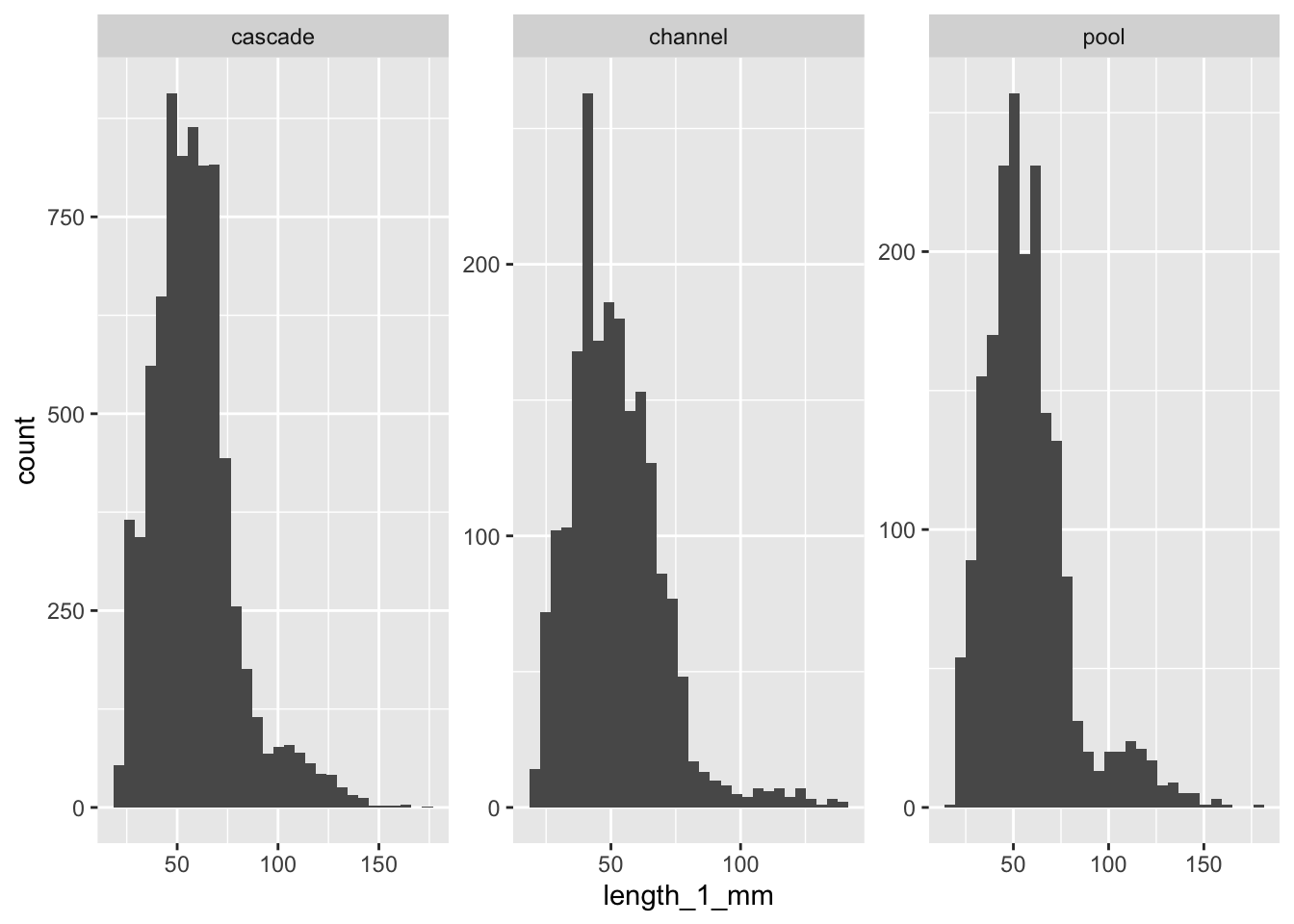
mutate(tf = log(length_1_mm))# A tibble: 3 × 6
unit_name mean sd count se var
<chr> <dbl> <dbl> <int> <dbl> <dbl>
1 cascade 58.3 20.8 7697 0.238 435.
2 channel 51.8 18.0 1994 0.403 324.
3 pool 57.3 22.8 1943 0.517 520.Unit name | Mean length (mm) | Standard deviation | Number of observations | Standard error | Variance |
|---|---|---|---|---|---|
cascade | 58.31985 | 20.84792 | 7,697 | 0.2376304 | 434.6357 |
channel | 51.77432 | 18.00636 | 1,994 | 0.4032398 | 324.2290 |
pool | 57.28358 | 22.80015 | 1,943 | 0.5172510 | 519.8469 |
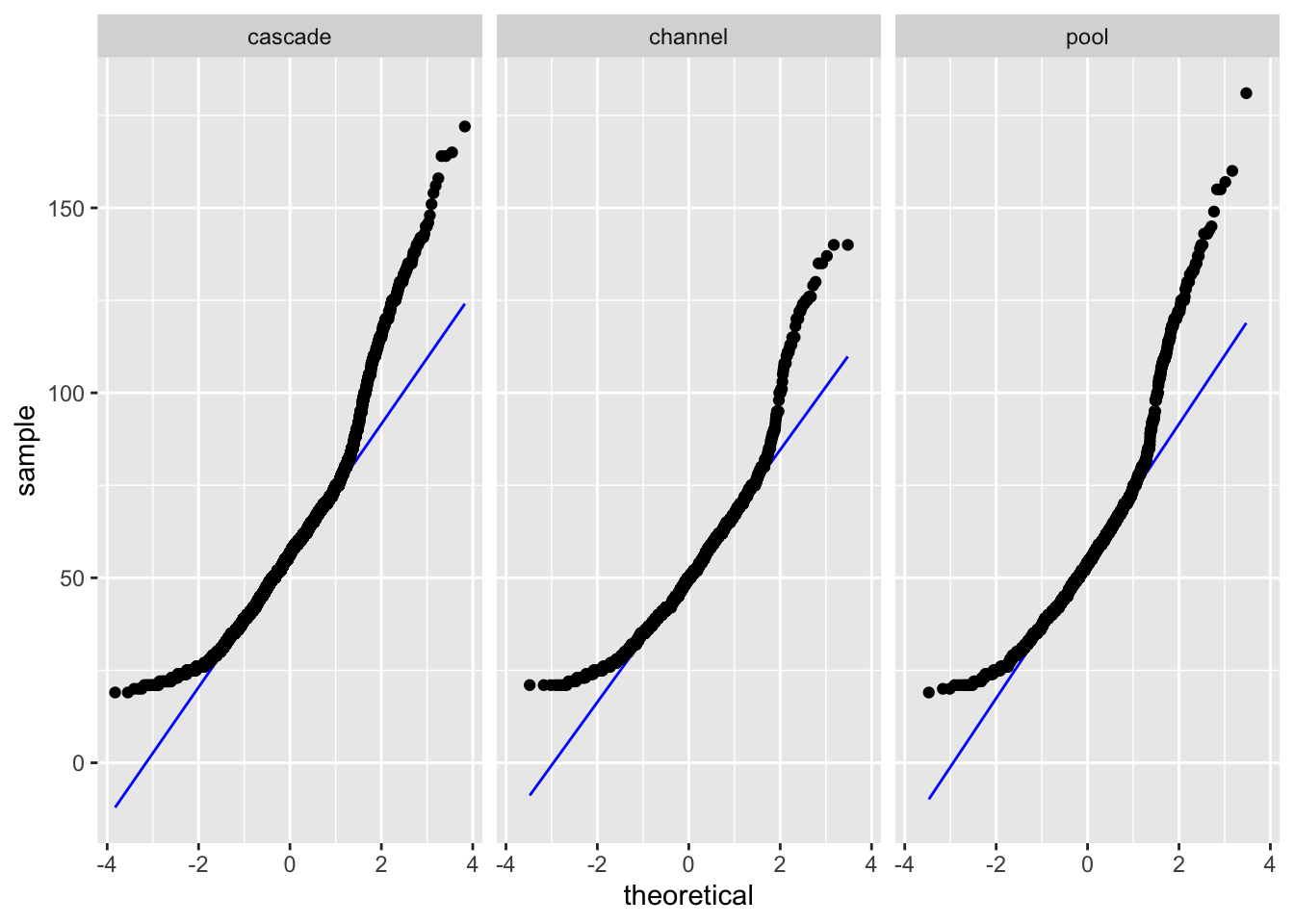
Levene's Test for Homogeneity of Variance (center = median)
Df F value Pr(>F)
group 2 20.922 8.516e-10 ***
11622
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Df Sum Sq Mean Sq F value Pr(>F)
unit_name 2 67994 33997 79.07 <2e-16 ***
Residuals 11622 4996776 430
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
9 observations deleted due to missingness Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = length_1_mm ~ unit_name, data = sal)
$unit_name
diff lwr upr p adj
channel-cascade -6.545526 -7.766976 -5.3240763 0.0000000
pool-cascade -1.036267 -2.270380 0.1978461 0.1201769
pool-channel 5.509259 3.959921 7.0585969 0.0000000tables:
(if we have time)
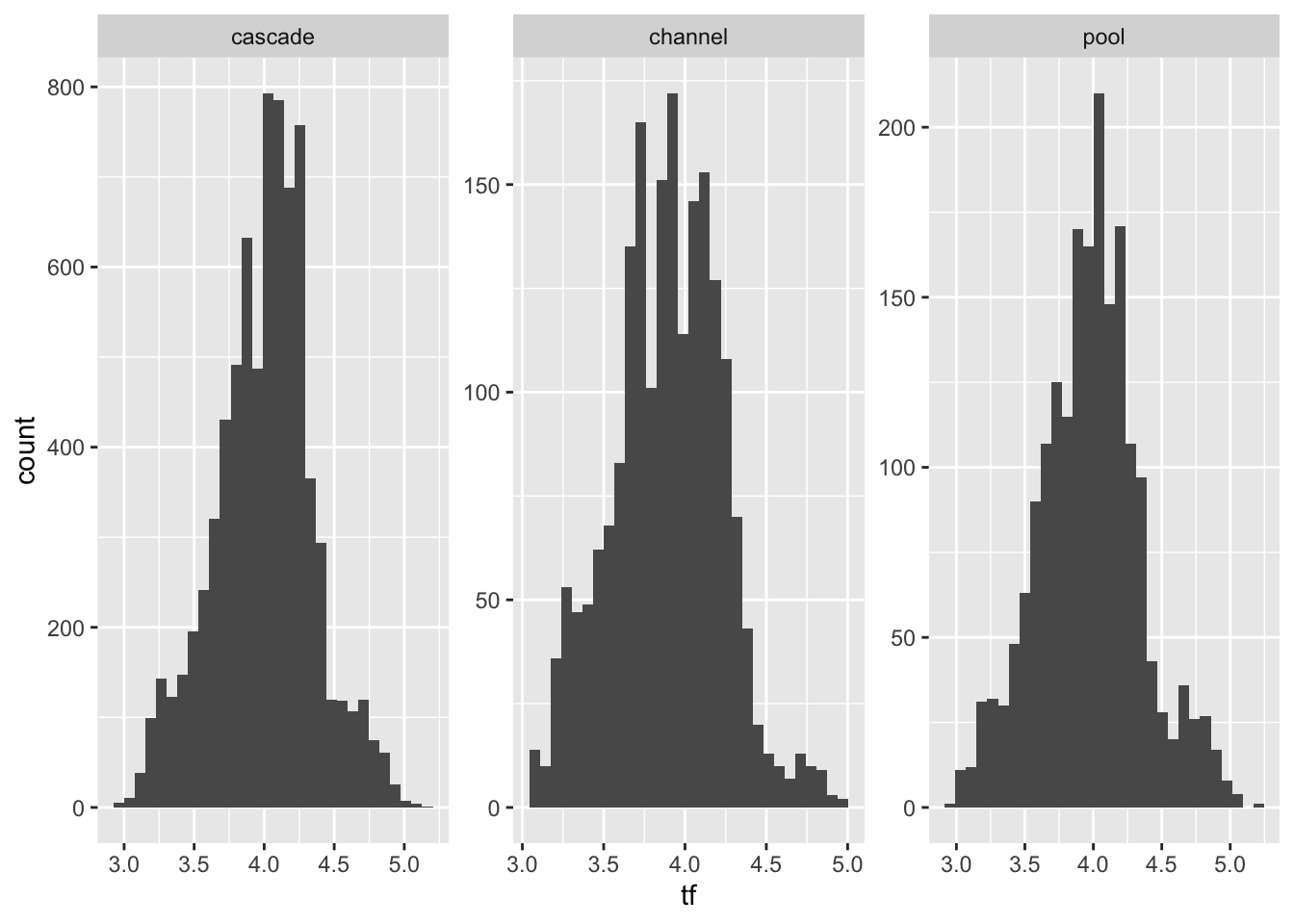
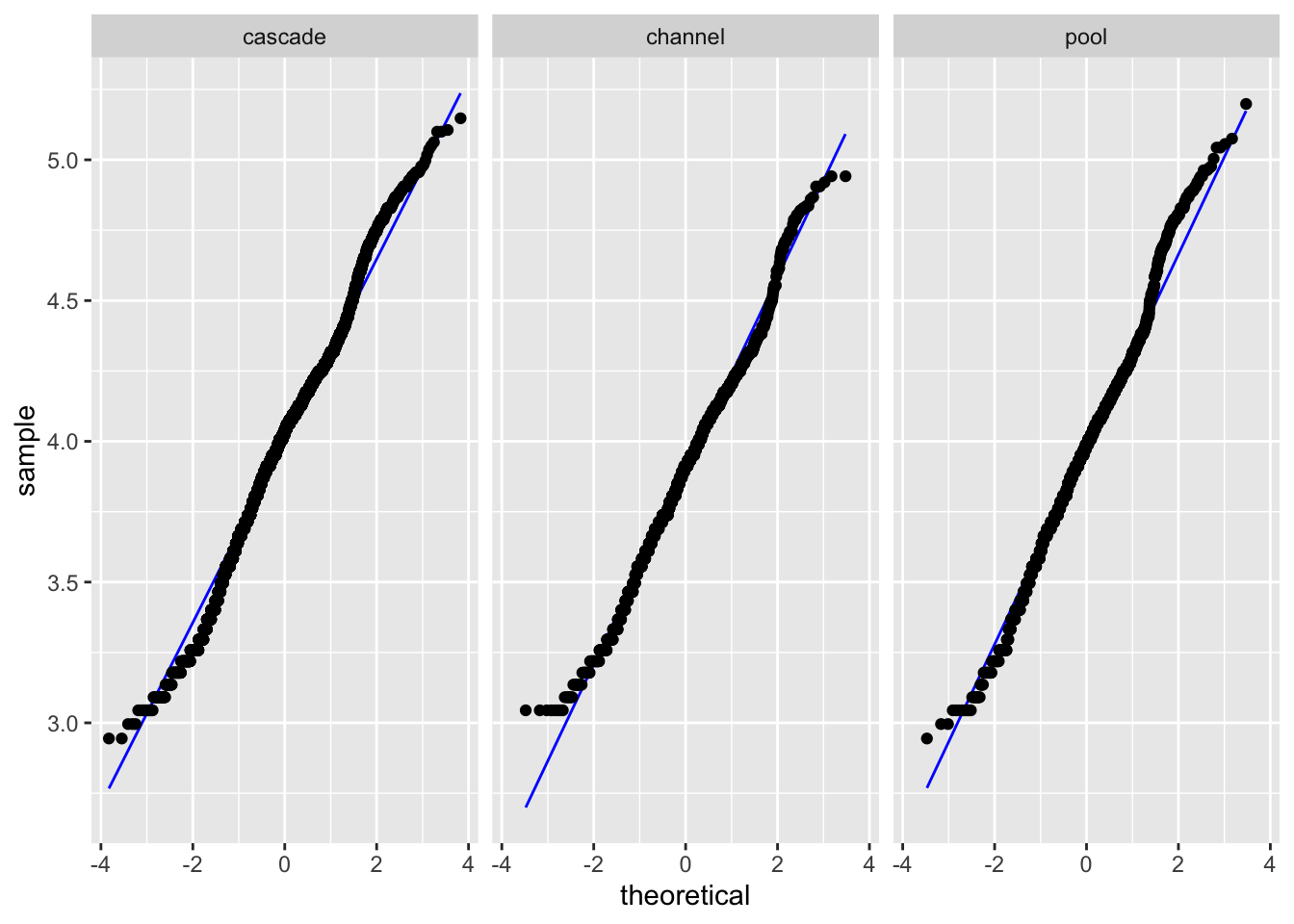
Levene's Test for Homogeneity of Variance (center = median)
Df F value Pr(>F)
group 2 6.103 0.002243 **
11622
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Df Sum Sq Mean Sq F value Pr(>F)
unit_name 2 20.9 10.431 83.95 <2e-16 ***
Residuals 11622 1444.1 0.124
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
9 observations deleted due to missingness Tukey multiple comparisons of means
95% family-wise confidence level
Fit: aov(formula = tf ~ unit_name, data = sal)
$unit_name
diff lwr upr p adj
channel-cascade -0.11470205 -0.13546713 -0.093936962 0.0000000
pool-cascade -0.02803709 -0.04901746 -0.007056729 0.0049454
pool-channel 0.08666495 0.06032566 0.113004248 0.0000000@online{bui2023,
author = {Bui, An},
title = {Coding Workshop: {Week} 7},
date = {2023-05-17},
url = {https://an-bui.github.io/ES-193DS-W23/workshop/workshop-07_2023-05-17.html},
langid = {en}
}